Aquatic ectotherms are predicted to harbour genomic signals of local adaptation resulting from selective pressures driven by the strong influence of climate conditions on body temperature. We investigated local adaptation in redband trout (Oncorhynchus mykiss gairdneri) using genome scans for 547 samples from 11 populations across a wide range of habitats and thermal gradients in the interior Columbia River. We estimated allele frequencies for millions of single nucleotide polymorphism loci (SNPs) across populations using low-coverage whole genome resequencing, and used population structure outlier analyses to identify genomic regions under divergent selection between populations. Twelve genomic regions showed signatures of local adaptation, including two regions associated with genes known to influence migration and developmental timing in salmonids (GREB1L, ROCK1, SIX6). Genotype–environment association analyses indicated that diurnal temperature variation was a strong driver of local adaptation, with signatures of selection driven primarily by divergence of two populations in the northern extreme of the subspecies range. We also found evidence for adaptive differences between high-elevation desert vs. montane habitats at a smaller geographical scale. Finally, we estimated vulnerability of redband trout to future climate change using ecological niche modelling and genetic offset analyses under two climate change scenarios. These analyses predicted substantial habitat loss and strong genetic shifts necessary for adaptation to future habitats, with the greatest vulnerability predicted for high-elevation desert populations. Our results provide new insight into the complexity of local adaptation in salmonids, and important predictions regarding future responses of redband trout to climate change.
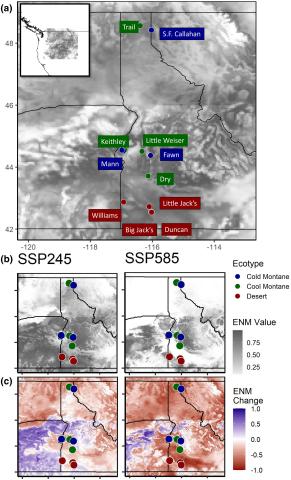
Map of study sites and ecological niche model (ENM) results for the present day, and for the period 2081–2100 across two shared-socioeconomic pathways (SSP245 and SSP585). a) ENM values for the present day, with higher ENM values representing higher relative probability of presence (a proxy for habitat quality). b) Raw ENM value for future time period. c) Change from present day ENM values for future time period. “Cold” = cold montane forest, “Cool” = cool montane forest, “Desert” = high-elevation desert
| GEM3 author(s) | |
| Year published |
2022
|
| Journal |
Molecular Ecology
|
| DOI/URL | |
| Keywords |
Bioinformatics
Ecology
Genomics
Redband Trout
|
| GEM3 component |
Mechanisms
|
| Mentions grant |
Yes
|