Severe drought conditions and extreme weather events are increasing worldwide with climate change, threatening the persistence of native plant communities and ecosystems. Many studies have investigated the genomic basis of plant responses to drought. However, the extent of this research throughout the plant kingdom is unclear, particularly among species critical for the sustainability of natural ecosystems. This study aimed to broaden our understanding of genome-to-phenome (G2P) connections in drought-stressed plants and identify focal taxa for future research. Bioinformatics pipelines were developed to mine and link information from databases and abstracts from 7730 publications. This approach identified 1634 genes involved in drought responses among 497 plant taxa. Most (83.30%) of these species have been classified for human use, and most G2P interactions have been described within model organisms or crop species. Our analysis identifies several gaps in G2P research literature and database connectivity, with 21% of abstracts being linked to gene and taxonomy data in NCBI. Abstract text mining was more successful at identifying potential G2P pathways, with 34% of abstracts containing gene, taxa, and phenotype information. Expanding G2P studies to include non-model plants, especially those that are adapted to drought stress, will help advance our understanding of drought responsive G2P pathways.
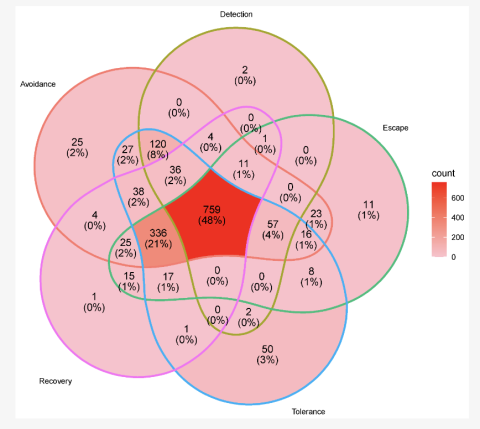
Venn diagram of the five major drought response strategies (avoidance, detection, escape, tolerance, and recovery) showing the overlap of genes associated with each category.
| GEM3 author(s) | |
| Year published |
2022
|
| Journal |
International Journal of Molecular Sciences
|
| DOI/URL | |
| Keywords |
Bioinformatics
Ecology
Genomics
Geography
Physiology
Genotyping by Sequencing
Functional Genomics
Landscapes
Sagebrush
|
| GEM3 component |
Mechanisms
|
| Mentions grant |
Yes
|