The quantity and nutritional quality of forage are key drivers for ungulate populations, including mule deer (Odocoileus hemionus) and Rocky Mountain elk (Cervus elaphus nelsoni), in the western U.S., but current vegetation maps are too coarse spatially and temporally to effectively characterize fine-scale habitat. To address some of these gaps, we tested a novel approach using existing vegetation surveys, maps, and remotely sensed data to develop fine-scale forage species distribution models (SDMs) across Idaho, USA. We modelled 20 forage species that are suitable for mule deer and Rocky Mountain elk. Climatic, topographic, soil, vegetation, and disturbance variables were attributed to approximately 44.3 million habitat patches generated using multi-scale object-oriented image analysis. Lasso logistic regression was implemented to produce predictive SDMs. We evaluated if the inclusion of distal environmental variables (i.e., indirect effects) improved model performance beyond the inclusion of proximal variables (i.e., direct physiological effect) only. Our results showed that all models provided higher predictive accuracy than chance, with an average AUC across the 20 forage species of 0.84 for distal and proximal variables and 0.81 for proximal variables only. This indicated that the addition of distal variables improved model performance. We validated the models using two independent datasets from two regions of Idaho. We found that predicted forage species occurrence was on average within 10% of observed occurrence at both sites. However, predicted occurrences had much less variability between habitat patches than the validation data, implying that the models did not fully capture fine-scale heterogeneity. We suggest that future efforts will benefit from additional fine resolution (i.e., less than 30 m) environmental predictor variables and greater accounting of environmental disturbances (i.e., wildfire, grazing) in the training data. Our approach was novel both in methodology and spatial scale (i.e., resolution and extent). Our models can inform ungulate nutrition by predicting the occurrence of forage species and aide habitat management strategies to improve nutritional quality.
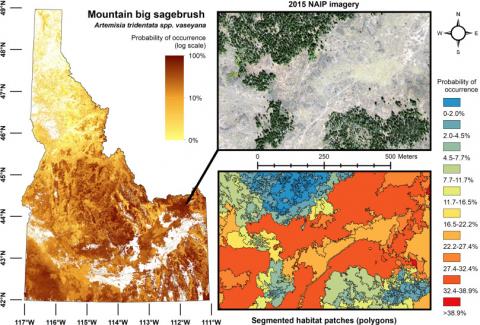
Habitat patches (i.e., polygons) developed from NAIP imagery and attributed with percent probability occurrence of mountain big sagebrush using a model containing distal and proximal (distal-proximal) environmental variables. Statewide values are mapped using a natural log scale and aggregated to 30 m resolution. The inset example is from the Caribou-Targhee National Forest in eastern Idaho.
| GEM3 author(s) | |
| Year published |
2020
|
| Journal |
Ecological Informatics
|
| DOI/URL | |
| GEM3 component |
Mapping
Modeling
|
| Mentions grant |
Yes
|