Current and past climatic changes can shift plant climatic niches, which may cause spatial overlap or separation between related taxa. The former often leads to hybridization and introgression, which may generate novel variation and influence the adaptive capacity of plants. An additional mechanism facilitating adaptations to novel environments and an important evolutionary driver in plants is polyploidy as the result of whole genome duplication. Artemisia tridentata (big sagebrush) is a landscape-dominating foundational shrub in the western United States which occupies distinct ecological niches, exhibiting diploid and tetraploid cytotypes. Tetraploids have a large impact on the species’ landscape dominance as they occupy a preponderance of the arid spectrum of A. tridentata range. Three distinct subspecies are recognized, which co-occur in ecotones – the transition zone between two or more distinct ecological niches – allowing for hybridization and introgression. Here we assess the genomic distinctiveness and extent of hybridization among subspecies at different ploidies under both contemporary and predicted future climates. We sampled five transects throughout the western United States where a subspecies overlap was predicted using subspecies-specific climate niche models. Along each transect, we sampled multiple plots representing the parental and the potential hybrid habitats. We performed reduced representation sequencing and processed the data using a ploidy-informed genotyping approach. Population genomic analyses revealed distinct diploid subspecies and at least two distinct tetraploid gene pools, indicating independent origins of the tetraploid populations. We detected low levels of hybridization (2.5%) between the diploid subspecies, while we found evidence for increased admixture between ploidy levels (18%), indicating hybridization has an important role in the formation of tetraploids. Our analyses highlight the importance of subspecies co-occurrence within these ecotones to maintain gene exchange and potential formation of tetraploid populations. Genomic confirmations of subspecies in the ecotones support the subspecies overlap predicted by the contemporary climate niche models. However, future mid-century projections of subspecies niches predict a substantial loss in range and subspecies overlap. Thus, reductions in hybridization potential could affect new recruitment of genetically variable tetraploids that are vital to this species’ ecological role. Our results underscore the importance of ecotone conservation and restoration.
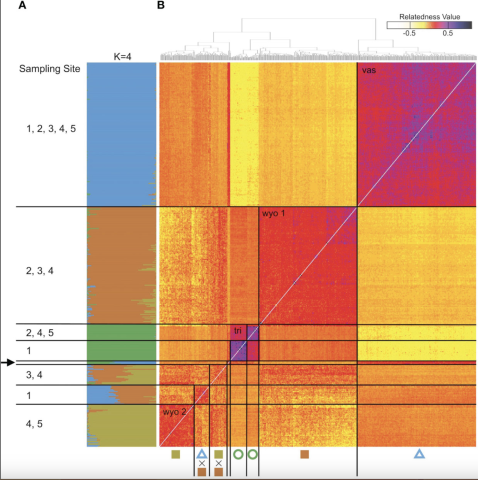
Genomic variation in A. tridentata. (A) Ancestry proportions bar plot as identified with STRUCTURE based on 540 SNPs and the four-population model (K=4). Each bar corresponds to an individual and each color to a genetic cluster. Individuals are sorted by the dendrogram inferred by the relatedness heatmap. (B) Pairwise relatedness heatmap based on 1,266 SNPs between 434 heteroploid individuals from 21 plots across the western United States. Numbers on the left indicate transect sites contained in a group, the arrow indicates the group containing diploid hybrids from various sites. The legend on the top right corresponds to the relatedness coefficients as estimated by Ritland’s method-of-moments estimator, the diagonal (i.e., individual compared to itself) was removed for improved contrast. Symbols below correspond to Figure 1.
| GEM3 author(s) | |
| Year published |
2023
|
| Journal |
Frontiers in Plant Science
|
| DOI/URL | |
| Keywords |
Bioinformatics
Ecology
Genomics
Genotyping by Sequencing
Population Genetics
Sagebrush
|
| GEM3 component |
Mechanisms
|
| Mentions grant |
Yes
|