Sage-grouse are two closely related iconic species of the North American West, with historically broad distributions across sagebrush-steppe habitat. Both species are dietary specialists on sagebrush during winter, with presumed adaptations to tolerate the high concentrations of toxic secondary metabolites that function as plant chemical defenses. Marked range contraction and declining population sizes since European settlement have motivated efforts to identify distinct population genetic variation, particularly that which might be associated with local genetic adaptation and dietary specialization of sage-grouse. We assembled a reference genome and performed whole-genome sequencing across sage-grouse from six populations, encompassing both species and including several populations on the periphery of the species ranges. Population genomic analyses reaffirmed genome-wide differentiation between greater and Gunnison sage-grouse, revealed pronounced intraspecific population structure, and highlighted important differentiation of a small isolated population of greater sage-grouse in the northwest of the range. Patterns of genome-wide differentiation were largely consistent with a hypothesized role of genetic drift due to limited gene flow among populations. Inferred ancient population demography suggested persistent declines in effective population sizes that have likely contributed to differentiation within and among species. Several genomic regions with single-nucleotide polymorphisms exhibiting extreme population differentiation were associated with candidate genes linked to metabolism of xenobiotic compounds. In vitro activity of enzymes isolated from sage-grouse livers supported a role for these genes in detoxification of sagebrush, suggesting that the observed interpopulation variation may underlie important local dietary adaptations, warranting close consideration for conservation strategies that link sage-grouse to the chemistry of local sagebrush.
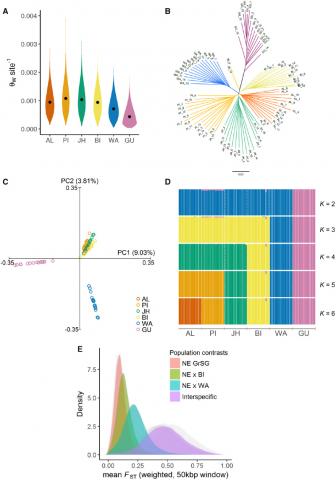
Figure 2. Inter- and intra-specific population genomic analyses of sage-grouse based on whole-genome resequencing. (A) Violin plots of mean per-site autosomal nucleotide diversity (Watterson’s estimator, θW) across populations. Values were estimated from 50-kb sliding windows. Median values are plotted within each kernel density curve. (B) Neighbor-joining tree based on pairwise genetic distances at autosomal SNPs, with branches colored by population. (C) Principal components analysis of complete data set, based on 1,500,781 nuclear SNPs. Axes represent first (PC1) and second (PC2) principal components, with percentage of total genetic variance explained by each component shown in parentheses.
| GEM3 author(s) | |
| Year published |
2019
|
| Journal |
Genome Biology and Evolution
|
| DOI/URL | |
| Keywords |
Ecology
Evolution
Genomics
Sagebrush
|
| GEM3 component |
Mechanisms
|
| Mentions grant |
Yes
|